Patsnap’s Biomedical AI Paper Recognized at EMNLP 2025
Currently Playing Post:Patsnap’s Biomedical AI Paper Recognized at EMNLP 2025
Patsnap is proud to share that our paper, “C.R.A.B: A Benchmark for Evaluating Curation of Retrieval-Augmented LLMs in Biomedicine,” has been accepted at EMNLP 2025, one of the world’s leading conferences in natural language processing.
As the first multilingual benchmark designed to evaluate how retrieval-augmented large language models (LLMs) curate biomedical content, this recognition marks a key milestone for Patsnap’s AI research — and a strong validation of our work at the intersection of artificial intelligence and biomedicine.
Access the paper here.
What Is C.R.A.B?
Patsnap’s paper introduces C.R.A.B, the first multilingual benchmark for evaluating how well retrieval-augmented LLMs curate biomedical content across English, French, German, and Chinese. Focused on a model’s “curation ability”, its skill in citing relevant sources and filtering out noise, C.R.A.B uses a novel citation-based metric to measure accuracy and efficiency. The results reveal clear performance gaps among leading models, highlighting the urgent need to strengthen biomedical curation capabilities.
Part 1: Evaluation of “Curation Ability”
As retrieval-augmented generation (RAG) gains traction in biomedical applications, reliably evaluating a model’s ability to curate relevant evidence has become a key challenge.
This refers to how well an LLM can identify and reference relevant literature while ignoring irrelevant materials within a retrieved corpus.
Unlike existing benchmarks, C.R.A.B introduces an innovative citation-based evaluation approach, assessing models based on their actual citation behavior during response generation based on three key metrics:
- Relevance Precision (RP): Accuracy of citing relevant evidence
- Irrelevance Suppression (IS): Effectiveness in avoiding irrelevant evidence
- Curation Efficiency (CE): Overall balance and robustness (reported as an F1 score)
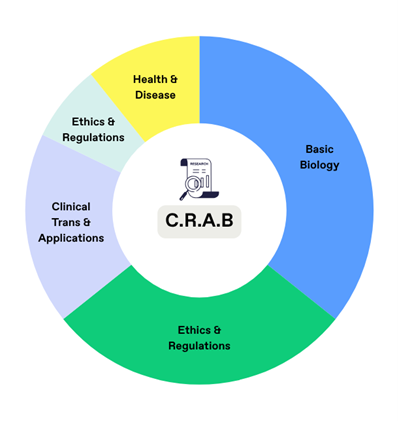
C.R.A.B features approximately 400 open-ended biomedical questions, paired with 2,467 relevant references and 1,854 irrelevant references covering topics such as basic biology, drug discovery, clinical applications, ethics, regulation, and public health. Some questions intentionally lack valid references to simulate real-world retrieval gaps.
All data were sourced from PubMed and Google Search, with retrieval powered by LlamaIndex.
Part 2: Key Findings
Patsnap evaluated 10 leading general and reasoning LLMs, including GPT-4-turbo, GPT-4o, Claude-3.7-Sonnet, Gemini-1.5-Pro, DeepSeek-V3, Doubao, DeepSeek-R1, QwQ-32B, Gemini-2.0-Thinking, and o3-mini, and recorded key findings below.
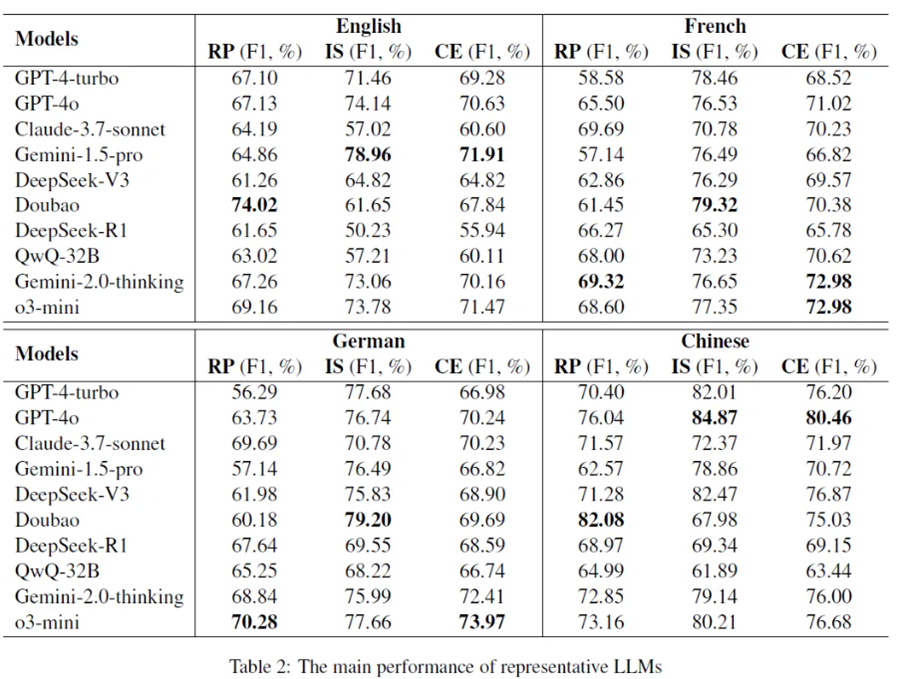
1st Finding: Closed-source models deliver more stable performance
Closed-source LLMs demonstrated stronger and more consistent performance.
- GPT-4o achieved the best results in Chinese (CE F1 = 80.46)
- Gemini-1.5-Pro led in English (CE F1 = 71.91)
- o3-mini excelled in French (72.98) and German (73.97)
This suggests that large, multilingual pretraining gives closed models a stability advantage over open-source ones.
2nd Finding: Some reasoning models underperform their base versions
Surprisingly, certain reasoning-focused models (e.g., DeepSeek-R1, QwQ) underperformed compared to their base counterparts. The research found that reinforcement training on math and code tasks led to “overthinking” in biomedical contexts, resulting in misuse of references and reduced domain accuracy.
3rd Finding: Domain-specific training significantly boosts performance
To test improvement potential, Patsnap performed continued pretraining (CPT) and supervised fine-tuning (SFT) on Llama 3-70B using biomedical data.
- Baseline CE F1 = 58.42
- CPT improved to 69.23
- SFT further raised it to 73.58
This proves that targeted domain training can markedly enhance evidence curation ability.
In addition, human expert validation on the English dataset confirmed C.R.A.B’s reliability: its automated evaluations matched expert judgments with over 90% consistency across all three metrics (RP 92.3%, IS 89.7%, CE 91.5%).
Accurate citation in biomedicine directly impacts clinical decisions, drug development, and patient safety. By quantifying LLMs’ evidence curation performance, C.R.A.B provides a unified standard for building trustworthy and transparent biomedical AI systems which proves valuable to clinical decision support, drug information retrieval, literature reviews, regulatory compliance, and more.
The CRAB dataset is now available on Hugging Face.
Looking Ahead
Patsnap remains committed to empowering innovation with cutting-edge AI.
This milestone in biomedical AI research not only enhances the quality of retrieval-augmented LLM applications in specialized domains but also strengthens the technical foundation of Patsnap’s AI products, helping customers extract deeper insights and make faster, more confident decisions across massive biomedical datasets. As Patsnap continues to push the boundaries of technology and product experience, it will remain a key driving force in enabling scientific and technological innovation worldwide.